
On the other hand, the DNA Pol α/primase complex places frequent RNA primers, 7–14 nucleotides (nt) in length, on the lagging strand and elongates them by addition of 10–20 deoxyribonucleotides. As a result, a highly processive polymerization complex is assembled, which carries out the leading strand DNA synthesis ( Lovett, 2007 Kunkel and Burgers, 2008 Burgers, 2009). On the leading strand, Pol α is displaced by the combined action of replication factor C (RFC), proliferating cell nuclear antigen (PCNA) and Pol ɛ ( Lovett, 2007 Kunkel and Burgers, 2008 Burgers, 2009). Primase synthesizes primers to initiate both leading and lagging strand synthesis.
#Okazaki fragment quizlet series
To achieve this, replication is semi-discontinuous: the leading strand is synthesized continuously by polymerase ε (Pol ε), from a single initiation event at the replication origin, whereas the lagging strand is initiated by the primase and is extended by polymerase δ (Pol δ) as a series of discrete Okazaki fragments ( Pursell et al., 2007 Kunkel and Burgers, 2008 Nick McElhinny et al., 2008 Burgers, 2009). As the replication machinery moves along the DNA, the synthesis of both new strands occurs simultaneously.

In eukaryotic cells, DNA replication initiates at multiple origins on each chromosome and proceeds bi-directionally from each origin into the flanking DNA, forming a DNA replication fork (Figure 1). Replication of double-stranded DNA is the central process in cell proliferation. Okazaki fragment maturation, FEN1, DNA2, PCNA, RPA, cancer Introduction On the other hand, mutations that impair editing out of polymerase α incorporation errors result in cancer displaying a strong mutator phenotype. These DNA strand breaks can cause varying forms of chromosome aberrations, contributing to development of cancer that associates with aneuploidy and gross chromosomal rearrangement. Mutations that affect the efficiency of RNA primer removal may result in accumulation of unligated nicks and DNA double-strand breaks. We also discuss studies using mutant mouse models that suggest two distinct cancer etiological mechanisms arising from defects in different steps of Okazaki fragment maturation. Such protein–protein interactions may be regulated by post-translational modifications. The dynamic interactions of polymerase δ, FEN1 and DNA ligase I with proliferating cell nuclear antigen allow these enzymes to act sequentially during Okazaki fragment maturation. Recent findings reveal that Okazaki fragment maturation is highly coordinated. Here, we summarize the distinct roles of these nucleases in different pathways for removal of RNA/DNA primers. The processing of RNA/DNA primers requires a group of structure-specific nucleases typified by flap endonuclease 1 (FEN1). During this process, primase-synthesized RNA/DNA primers are removed, and Okazaki fragments are joined into an intact lagging strand DNA. Therefore, efficient processing of Okazaki fragments is vital for DNA replication and cell proliferation. Even in yeast, the Okazaki fragment maturation happens approximately a million times during a single round of DNA replication.
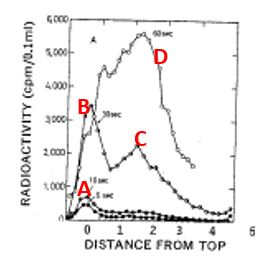
Completion of lagging strand DNA synthesis requires processing of up to 50 million Okazaki fragments per cell cycle in mammalian cells.


 0 kommentar(er)
0 kommentar(er)
